Characterizing the Microbiome of Dysbiosis
Disturbances of the intestinal microorganisms can compromise the health of the host. Persistent imbalanced states that lead to disease are referred to as dysbioses. Several patterns of dysbiosis have been associated with particular diseases. However, we are still a long way from being able to definitely conclude that certain patterns of bacteria are always (or often) associated with a particular condition. Our research focus is to characterize these patterns from a variety of diseases to understand which features might be mechanistically linked to a particular pathology. A central theme in our group is finding ways to define resilience among the intestinal microbiota and testing these interpretations to see if they are informative of outcomes.
Research Projects
We collaborate with a wide variety of highly motivated physician and basic scientists, and our on-going research projects include:
Human-Microbiota Associated Model
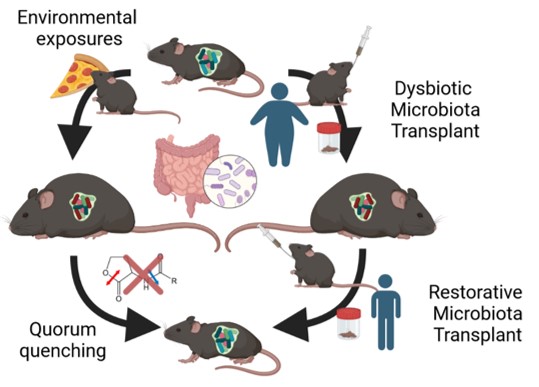
- Improve transplantation of human microbiota into antibiotic-conditioned, conventionally housed mice
- Determine causal pathological mechanisms of the microbiota using human microbiota-associated mouse model
The Microbiome in Surgical Healing
- Characterizing changes in the microbiome associated with surgery and peri-operative antibiotics
- Determining mechanistic roles of the microbiota in surgical outcomes using mouse models
Quorum Quenching Studies
- Evaluate the in vivo effects of quorum quenching lactonases on microbiota composition and function
- Determine efficacy of quorum quenching to mitigate pathological processes